Digital PCR Assay Optimization
Sub-optimal digital PCR settings very often lead to insufficient signal to noise ratio. This affects the separation between the negative and the positive partitions and prevents for an adequate threshold setting. That leads to limit the accuracy of the quantification and the sensitivity of the assay.
When developing an assay on a digital PCR system, there are several elements to consider:
- DNA template for assay optimization (positive control)
- Difference in fluorescence amplitude between negative and positive populations
- Presence of partitions of intermediate fluorescence (rain)
- Specificity – Are there non-specific populations? Can you detect positive partitions in your negative control?
1. DNA template for assay optimization
Ideally, the DNA template should be in the same form as it will be in the sample you will want to assay : sheared in small fragments if targeting circulating cell-free DNA or relatively intact if assaying genomic DNA. The DNA solution used should be devoid of contaminants and potential inhibitors and have good absorbance (A260/230 and A230/260) ratios. If there is no evident source for DNA material to develop your assay, you may use synthetic oligos as templates for optimization.
-
Presence of false positives
False positives are also a factor that contributes to the diminution of the sensitivity of the digital PCR assay. To limit this phenomenon, it is crucial to prevent DNA contamination in the laboratory or cross-contamination from well to well. Template quality is also a factor that is important to monitor as base degradation such as cytosine deamination and 8-Oxo-2′-deoxyguanosine formation due to oxidative damage may lead to base transversion during PCR amplification.
For more information about DNA preparation, have a look at the “DNA Preparation For Digital PCR” item.
2. Difference in fluorescence amplitude between negative and positive populations
When you run an assay for the first time, you might not distinguish the negative and positive populations on a digital PCR system (Figure A).
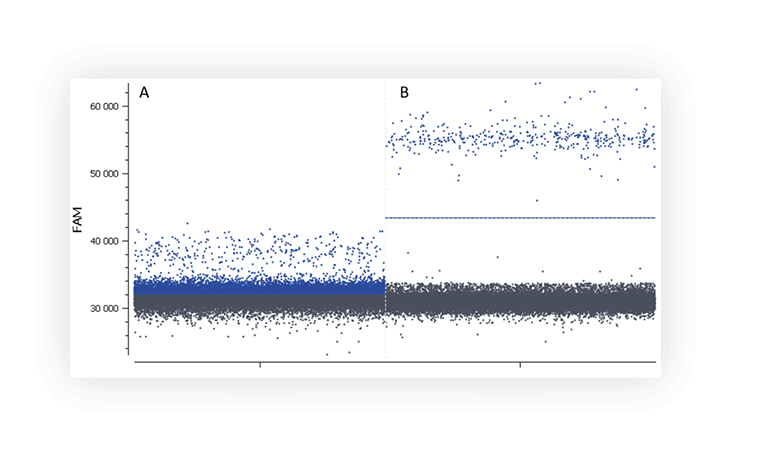
Figure A. The positive partition population (blue) is barely separated from the negative population (dark grey), rendering thresholding difficult on this 1D dot plot. B. The fluorescence of the positive population (blue) is clearly separated from the negative population (dark grey), placing a threshold is therefore unambiguous.
Here are a few tips to improve the separability of your populations:
- Check manufacturers recommendations on primers and probe concentrations to be used in your digital PCR system. These may be higher than the ones usually recommended in qPCR.
- Optimize hybridization temperature. Check for the highest hybridization temperature that gives you an optimal separability and no rain (see below).
- Check the type of probes you are using. Double-quenched probes will give you a lower basal fluorescence signal, and therefore a higher separability.
- Check whether your probes are too old! Depending on how long your probes have been stored, whether they have been stored properly, or whether they just encountered too many freeze-thaw cycles, it is possible that your probes are already hydrolyzed. In this case, you may see a high fluorescence basal level, and, since there are only a few intact probes for PCR, a low signal for the positive partitions.
- If using a system where you can vary acquisition parameters, you may also try to tune the exposure time
3. Presence of partitions of intermediate fluorescence (rain)
We call “rain” partitions that have intermediate fluorescence values between the positive and the negative populations. In these partitions, amplification/hybridization efficiency is sub-optimal. The rain makes the threshold setting harder, and thus may affect quantification.
Various factors may be at the origin of the “rain” such as, template degradation, PCR inhibitors, polymerase errors, primer dimer, target accessibility (Figure B) or non-homogenous distribution of fluorescence into partitions.
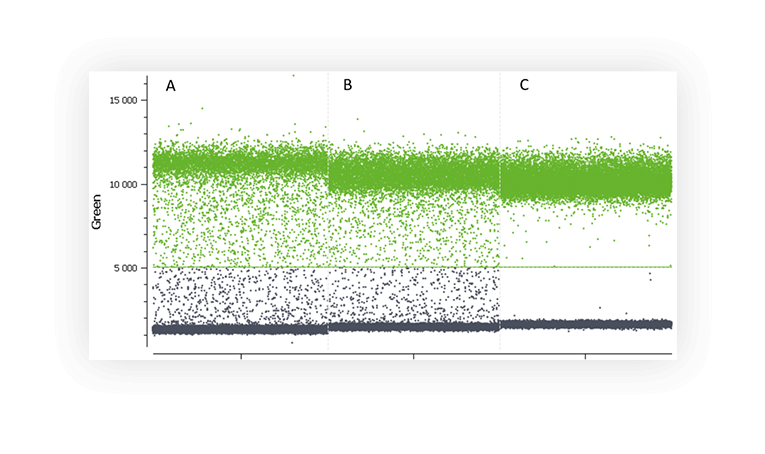
Figure B. 1D dot-plots of a digital PCR experiment in partition using one fluorescent assay targeting a non-digested plasmid (A and B) and a digested plasmid (C). Rain is clearly visible on the two first plot whereas plasmid digestion has removed most of the partitions corresponding to rain.
In order to minimize the rain, you can optimize your digital PCR reaction by:
- Checking the optimal hybridization temperature as described above
- Checking your DNA template for anything that may prevent accessibility to the target. Is this a GC-rich region? Try additives such as DMSO or betaine. When using high molecular-weight DNA or plasmids, it is also best to fragment the DNA by digestion or mechanical means prior to digital PCR. If using digestion, you may also be able to perform this step directly in the mix.
- Making sure to use DNA free from inhibitors
- Increasing the number of cycles in order to ensure that all partitions reach the reaction plateau.
4. Presence of non-expected populations
When assaying a single target, you usually expect a single positive population. But sometimes, you will discover that a second distinct population has appeared (Figure C). It can be due to primer dimers and non-specific amplification.
Multiple fluorescence populations may prevent from setting a proper threshold.
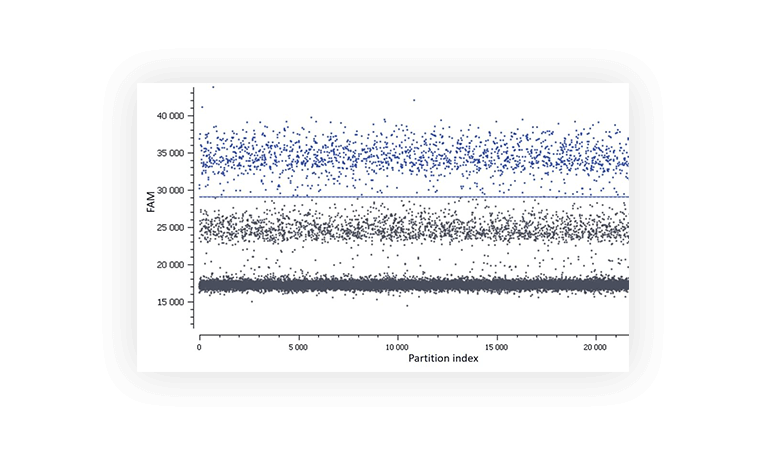
Figure C. In this assay, a second population is observed between the negative (lower grey band) and the positive (blue).
Various methods can be employed to limit this phenomenon such as optimization of primer design, increase of annealing temperature and touchdown PCR.
This specificity issue may be fixed:
- The hybridization temperature is potentially too low so make sure you pick the highest temperature that gives you optimal separability while minimizing the rain
- Have you thought about performing a touchdown PCR?
- Is this non-specific population really an issue ? In terms of quantification, you can just decide to place the threshold above it as we have done in Figure C. The non-specific population amplified will just be considered part of the negative population. The results will thus be calculated only for the amplified partitions that show a reaction efficiency consistent with a specific hybridization of the primers and probes. Please note that this is not the optimal way of analyzing your data since it can have an impact on precision and sensitivity.
- Re-design your primers. To do so, you may use online tools to check that your primers and probes do not hybridize elsewhere in the genome.
For more information on optimization to improve your digital PCR assay, see the following article:
Lievens, A., Jacchia, S., Kagkli, D., Savini, C., Querci, M. Measuring Digital PCR Quality: Performance Parameters and Their Optimization. PLoS One. 2016 May 5;11(5):e0153317. doi: 10.1371/journal.pone.0153317. eCollection 2016. PMID: 271494
